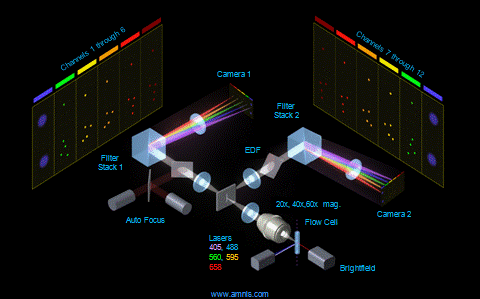
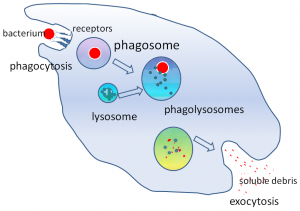
Phagosome maturation after bacterial uptake:
Dr. Katarzyna Jobin,
Scientist (Kurts Lab, Institute of experimental Immunology):
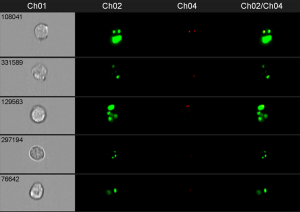
In this example phagosomes and lysosomes where stained in different colors. If both are colocalized in the cell, the phagosomes are considered as mature. Of course, the phagosomes could also be detected by fluorescence labeling of the bacteria prior the incubation with cells.
The power of this method, if conducted on the image stream, is that you can quantify the amount of mature and premature phagosomes and analyze them in different cell types (like neutrophils and macrophages) at the same time, without prior separation. You simply have to stain for the cell type specific surface makers to identify the different populations.
*Wikimedia Commons
Prime Flow:
Kasia also conducted the Prime Flow RNA Assay on the ImageStream.
Prime Flow RNA Assays are fluorescence in situ hybridization (FISH) assays that allow specific localization of RNA targets in flow. RNA detection can be combined with intracellular or surface stainings of proteins on a single cell level. PrimeFlow RNA Assay
These assays can also be measured in conventional flow cytometry, but in image cytometry you will get a better resolution, because the RNA signals are focused in discrete spots with rather low intensities. This kind of signals are hard to analyze in conventional cytometry because you only measure the overall fluorescence intensity of each cell, which might be just slightly above the background. To solve this problem, you can use image cytometry and then you are able to combine the fluorescence intensity and the localization of the signal.
Analysis of bacterial clearance combined with surface markers.
Victoria Scheiding,
PhD Student (Garbi Lab, Institute of experimental Immunology):
In Victorias project, she is interested in bacterial clearance in neutrophils. First experiments could already show, that the method is promising. The cells where stained with anti CD45 and anti Ly6G for identification and the bacteria express mCherry.
fig.1 fig.2
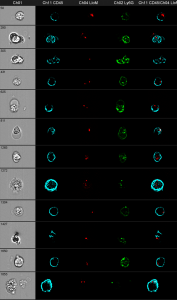
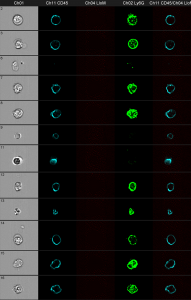 Gray column: brightfield image. The first black column shows the anti CD45 staining. The second black column shows the bacteria in red. The third one shows anti Ly6G and the fourth shows an overlay of all three fluorescence parameters.
Gray column: brightfield image. The first black column shows the anti CD45 staining. The second black column shows the bacteria in red. The third one shows anti Ly6G and the fourth shows an overlay of all three fluorescence parameters.
You can clearly distinguish between discrete red dots at the left figure compared to the wild-type control at the right. The next challenge is the optimization of the spot-counting algorithm to get precise results of the automated analysis.
Changes in mitochondrial function in HIV patients
Michael To Vinh,
PhD student (Nattermann Lab, Medizinische Klinik und Poliklinik I):
Michael wants to investigate mitochondrial differences in NK-cells between HIV patients and healthy donors. The samples were stained with surface markers like CD56 in order to gate on NK-cells and CD3 to exclude the NKT-cells. The mitochondria were stained with Mitotracker green. The amount of mitochondria per cell can be analyzed by spot count algorithms in high throughput on single cell level. This is more precise than standard flow cytometry, way faster and also less error-prone than microscopy. But as mentioned before, the automated analysis still needs some improvements.
![]()
First column: brightfield image; CH11: CD45; CH02: Mitotracker
ASC speck formation
Katarzyna Andryka
PhD student (Bartok Lab, Institut für Klinische Chemie und Klinische Pharmakologie)
In this experiment, Katarzyna tried to analyze the speck formation of the protein ASC during NLP3 inflamasome activation in M-CSF generated human macrophages. In a non activated state the protein is equally distributed within the whole cytoplasm of the cells. After inflamasome activation the ASC protein clusters in discrete regions of the cell and can be used as a reporter.

non specked
![]()
specked
Cell nucleus counting in cardiomyocytes
Cora Becker,
PhD student (Hesse Lab, Institut für Physiologie I):
Adult cardiomyocytes (CMs) of mammals have very often two nuclei. This makes cell cycle analysis and proliferation studies very difficult. The group of Dr. Michael Hesse has established a transgenic mouse model to visualize the CM nuclei in vitro and in vivo, by fusing the red fluorescent protein mCherry to human histone 2B (H2B) and expressing it in CMs by use of the αMHC (Myh6) promoter. Cora Becker used this reporter system now on the ImageStream system to benefit from its high throughput capabilities and the automated data analysis. Compared to conventional microscopy, this method is much faster and in combination with the autosampler it can even be used for screenings.
![]()
one nucleus
![]()
two nuclei