Our technologies
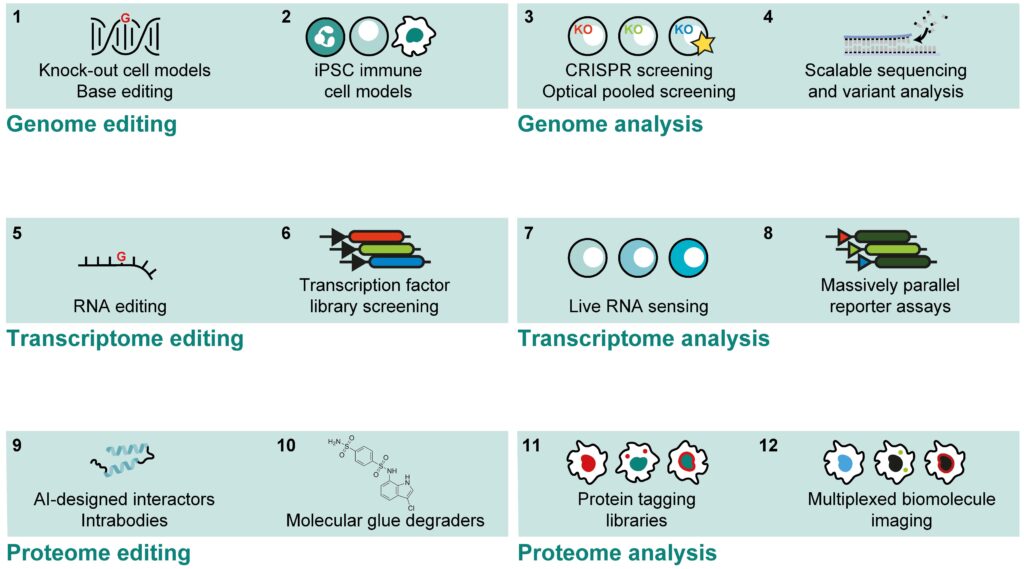
The Bonn Competence Hub for Functional Genomics uses and develops advanced techniques to delineate the molecular effects of specific genetic alterations affecting immune functions in the human population and identified in patients with suspected inborn errors of immunity (37084271, 39039281). Genetic variants are modelled in human immune cell types using a robotic CRISPR genome editing pipeline (25186908), which has enabled to understand immune gene functions in the past (24077100, 26174085). Molecular karyotyping, shallow genome sequencing or exome sequencing are used to ensure the identity and genome stability of cellular models. Beyond targeted genome editing, FACS-based (26553871) and Optical Pooled CRISPR Screening (31626775, https://doi.org/10.1101/2024.01.18.576210), highly parallel reporter assays (MPRA), and saturation mutagenesis screens are used to map the functional relevance of genetic factors, gene regulatory elements, or individual residues of immune-relevant genes and proteins. Researchers can interrogate the downstream functions of perturbed genes in model cells using well-established and novel omics methods (36138185, 34188222, https://doi.org/10.1101/2024.07.17.603943) and standardized sequencing data analysis to identify differentially regulated genes (26513548). Beyond modelling genetic alterations in cell lines, the Functional Genomics Hub bundles expertise to tag endogenous genes for live cell studies (27465542) and support well-established protocols for polyclonal genome editing in primary human T cells, PBMC-derived macrophages, and induced pluripotent stem cells (iPSC), which can be generated from patient-derived fibroblasts or obtained from a shared collection. Using large transcription factor libraries, new differentiation protocols have been established to obtain a variety of immune-relevant cell types (33257861). The competence hub also bundles expertise in generation, cultivation, genetic editing, and microscopic analysis of organoids derived from patient material or iPSCs to model organ physiology in three-dimensional cellular ensembles, and supports transgenic or gene-deficient mouse models. To probe for specific protein-protein interactions, post-translational modifications, or subcellular localizations affected by human genetic variants, intracellular nanobodies, AI-designed minibinders, and protein degraders are employed in highly parallelized experiments.
Our collaborations
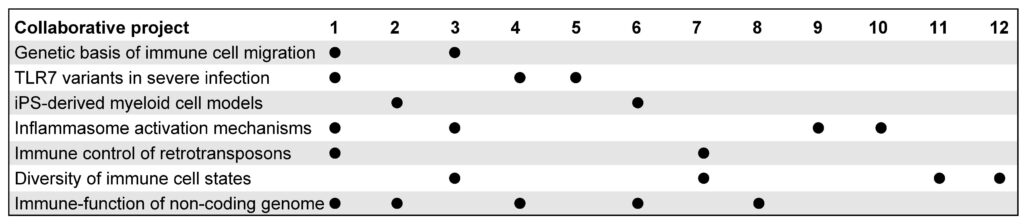
The competence hub will foster collaborative functional genomics projects across cluster groups. It will be coordinated by and integrated in the Schmid-Burgk lab and will bring together unique expertises, resources, and techniques across our campus. One full-time postdoctoral researcher and one full-time technician will develop, standardize, and apply novel functional genomics technologies, involved at the core of multiple cluster research projects (spatiotemporal dissection of innate signaling pathways, functional mapping of genetic variation in key immune genes, identification of defense genes by cross-species structural clustering, next generation CAR T and NK cells using orthogonal signaling pathways, designer immune modulators by engineering proximity of proteins, Statistics First immune data mining, identification of genetic circuits in immune cells by optical pooled screening, context-dependent analysis of functional effects of non-coding risk variants underlying severe COVID-19). Beyond basic funding, Cluster Flex Funds will be granted to collaborative projects in the area of functional analysis of immune-relevant genes, and a seminar series will bring together researchers applying and developing functional genomics techniques.
Steering Committee
Prof. Dr. Jonathan Schmid-Burgk
Prof. Dr. Florian Schmidt
Prof. Dr. Kerstin Ludwig
Dr. Anna Aschenbrenner
Dr. Lorenzo Bonaguro
Prof. Dr. Kaan Boztug
PD Dr. Michael Peitz
Prof. Dr. Michael Hölzel
Prof. Dr. Gregor Hagelüken
Prof. Dr. Maximilian Billmann
Prof. Dr. Hiroki Kato
Prof. Dr. Busskamp
Prof. Dr. Radoslaw Nowak
Prof. Dr. Joachim Schultze
Publications
- Ludwig KU, […], Hölzel M, […], Schmid-Burgk JL. LAMP-Seq enables sensitive, multiplexed COVID-19 diagnostics using molecular barcoding. Nature Biotechnology. 2021 Dec;39(12):1556-1562. doi: 10.1038/s41587-021-00966-9.
- Fandrey CI, Jentzsch M, […], Schmidt FI, […], Schmid-Burgk JL. NIS-Seq enables cell type agnostic optical perturbation screening. Nature Biotechnology, in press (2024).
- Rouillon C, […], Schmid-Burgk JL, […], Hagelueken G. Antiviral signaling by a cyclic nucleotide activated CRISPR protease. Nature. 2023 Feb;614(7946):168-174. doi: 10.1038/s41586-022-05571-7.
- Koenig PA, […], Schmid-Burgk JL, Kato H, […], Schmidt FI. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021 Feb 12;371(6530). doi: 10.1126/science.abe6230.
- Boos J,[…], Sacchi N. Spanish/Italian Severe COVID-19 Sequencing group; GenOMICC Investigators; Schultze JL, […], Ludwig KU. Stratified analyses refine association between TLR7 rare variants and severe COVID-19. HGG Adv. 2024 Oct 10;5(4):100323. doi: 10.1016/j.xhgg.2024.100323.
- Zieger HK, […], Ludwig KU. Prioritization of non-coding elements involved in non-syndromic cleft lip with/without cleft palate through genome-wide analysis of de novo mutations. HGG Adv. 2022 Dec 5;4(1):100166. doi: 10.1016/j.xhgg.2022.100166.
- COVID-19 Host Genetics Initiative. A second update on mapping the human genetic architecture of COVID-19. Nature. 2023 Sep;621(7977):E7-E26. doi: 10.1038/s41586-023-06355-3.
- Butler-Laporte G […]. COVID-19 Host Genetics Initiative; DeCOI Host Genetics Group; GEN-COVID Multicenter Study (Italy); Mount Sinai Clinical Intelligence Center; GEN-COVID consortium (Spain); GenOMICC Consortium; Japan COVID-19 Task Force; Regeneron Genetics Center; Geschwind DH, […] Ludwig KU, Exome-wide association study to identify rare variants influencing COVID-19 outcomes: Results from the Host Genetics Initiative. PLoS Genet. 2022 Nov 3;18(11):e1010367. doi: 10.1371/journal.pgen.1010367.
- Schmidt A, […], Ludwig KU. Predicting the pathogenicity of missense variants using features derived from AlphaFold2. Bioinformatics. 2023 May 4;39(5):btad280. doi: 10.1093/bioinformatics/btad280.
- Kircher M, Ludwig KU. Systematic assays and resources for the functional annotation of non-coding variants. Med Genet. 2022 Dec 31;34(4):275-286. doi: 10.1515/medgen-2022-2161.