Competence Hub Spatial Biology
Advances in next-generation sequencing and imaging-based approaches have led to a revolution in our ability to understand cellular function in spatial context.
The Competence Hub for Spatial Biology brings together expertise in method development, technology application, and mathematical analysis established by pioneering work in different research groups and institutes. By combining these resources, we can perform cutting-edge research in the fields of immunology, oncology, infectious diseases, and neurosciences.
Our aims are as follows:
- Access – bundle the instruments and know-how under an umbrella structure to provide access to spatial technologies across faculty(ies)
- Consulting – to enable researchers at the University to pick and use spatial biology and single cell technologies efficiently
- Tech-watch capabilities – to identify novel technological breakthroughs and stay ahead of the wave
- Interact – actively engage with leading companies within the spatial technology sector to foster co-developments
- Disseminate – Build a core facility for spatial computational analysis
Technologies
RNA
In situ capture (PRECISE)
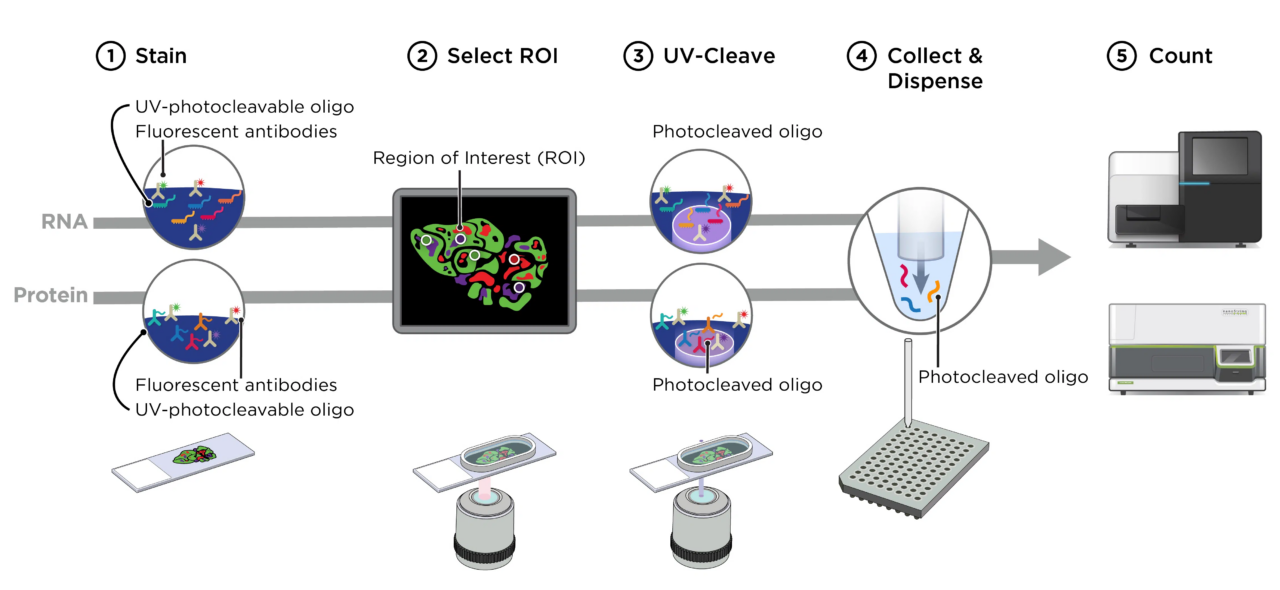
PRECISE operates a NanoString’s GeoMx Digital Spatial Profiler (DSP) which allows the user to define a microscopic region of interest on an FFPE or frozen tissue slide due to a UV-photocleavable barcode engineered into the in-situ hybridization probes (targeted or up to whole transcriptome level). The region of interest is specifically exposed to UV light, the barcodes are cleaved and used to identify the RNA or protein present in the tissue by next-generation sequencing. The defined regions of interest can vary in size between ten and six hundred micrometers allowing targeting of a wide variety of structures and cells in the histological sample with all downstream analysis steps fully established in PRECISE.
Marc Beyer (DZNE) marc.beyer@dzne.de
In situ sequencing with padlock probes (LIMES & PRECISE)
The Xenium spatial transcriptomics workflow with in situ sequencing (ISS) using padlock probes has been implemented at the LIMES and PRECISE. The ISS padlock method is based on padlock probing, rolling-circle amplification (RCA), and sequencing by ligation chemistry. Within intact tissue sections, mRNA is reversely transcribed to cDNA, which is followed by mRNA degradation by RNase H. Next, DNA circularization of a padlock probe with a barcode sequence is conducted by ligation only. Target amplification is then performed by RCA, yielding micrometer-sized RCA products (RCPs). RCAs consist of repeats of the padlock probe sequence. These DNA molecules are then subjected to sequencing by hybridization technology, decoding a several base-long barcode within the probe using microscopy equipment available at the LIMES and the DZNE. The further analytical workflow including imaging, preprocessing including segmentation and further bioinformatic analysis are established at the LIMES and the PRECISE.
Andreas Schlitzer (LIMES) andreas.schlitzer@uni-bonn.de
Marc Beyer (DZNE) marc.beyer@dzne.de
MERSCOPE Merfish
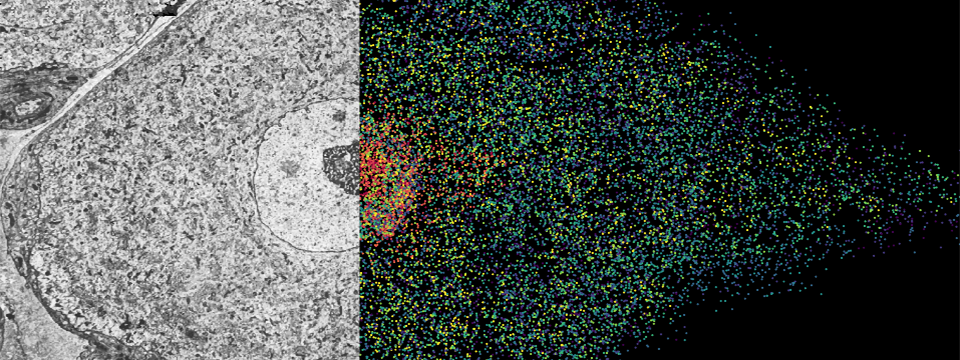
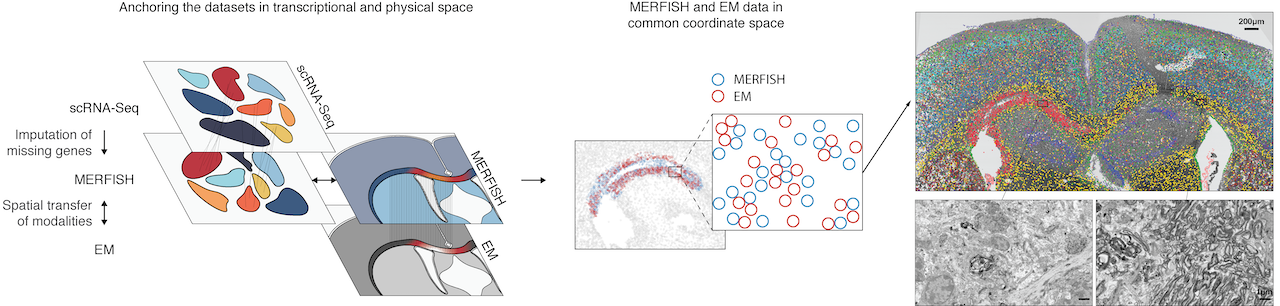
The MERFISH (Multiplexed Error-Robust Fluorescence in situ Hybridization) technology, a multiplexed imaging technique for identifying nucleic acids in their native tissue environment. MERFISH is capable of analyzing up to 10,000 RNA targets, as well as can be used to measure epigenetic changes in tissue. The Vizgen MERSCOPE is the commercial MERFISH Platform. Currently available kits can be applied to FFPE or frozen tissue sections upto 1000 RNA combined with 6 validated antibodies. Recently, the Gokce group has combined MERFISH with Electron Microscopy (Androvic et al., 2022) using adjacent tissue sections. This parallel analysis of adjacent tissue sections allows different omics technologies to be run in their optimal protocols and to be integrated computationally using spatial anchors.
Özgün Gökçe (DZNE, UKB) ozgun.goekce@ukbonn.de
Laser-capture microdissection-based OMICs (PRECISE)
LCM-OMICs utilizes laser capture microdissection (LCM) coupled with ATAC-seq or Smart-Seq2 RNA-seq and is applicable down to the single cell level and can even be used on partially degraded tissues to determine chromatin accessibility or gene expression. The workflow includes cryo-sectioning of tissues either directly (for RNA) or after Tn5 transposase treatment (for ATAC) followed by laser capture microdissection, where cells are collected directly into lysis buffer and cDNA is generated without the need for RNA resp. DNA isolation, which both simplifies the experimental procedures as well as lowers technical noise. As the positional identity of each cell is recorded during the LCM procedure, the chromatin state resp. the transcriptome of each cell after sequencing of the corresponding DNA library can be inferred to the position where it was isolated from. DZNE operates a Zeiss PALM microdissection setup with a robomover with automated artificial intelligence driven-object detection-delineation-cutting-and-collection workflow with all subsequent analysis steps including sequencing and bioinformatic analysis of LCM-based RNA-seq or ATAC-seq established at PRECISE.
Eugenio Fava (DZNE) Eugenio.Fava@dzne.de
Protein
Midiplex
A Midiplex system like the Axio Scan System from Zeiss allows to automatically acquire low- to medium-resolution images of tissue sections using 3-4 fluorescent markers. The ZEISS Axio Scan.Z1 slide scanner is available at the Light Microscopy Imaging Facility at the Medical Faculty and allows to digitize specimens and create high-quality virtual slides in a reliable, reproducible way. Furthermore, a custom Zeiss Axio Imager Z2 setup at UKB now allows to increase midiplexing up to 8 makers and to process 8 slides automatically. The DZNE also operates a Zeiss Axio Scan.Z1, a Zeiss Cell Discoverer7 including an LSM900 Airy Scan confocal for increased resolution (about 100nm), and a Zeiss PALM microdissection setup with a robomover with automated artificial intelligence driven-object detection-delineation-cutting-and-collection workflow.
Contact:
Hannes Beckert (Core Facility, UKB): hbeckert@uni-bonn.de
Eugenio Fava (DZNE) Eugenio.Fava@dzne.de
Highplex
Several high-plex imaging systems utilizing the Akoya CODEX Phenocycler platform coupled to Zeiss Axio Observer 7 exist at the Life & Medical Sciences Institute and the UKB. This system allows the simultaneous characterization of up to 192 protein markers on tissue sections with high-resolution imagery. The DZNE also operates an Akoya CODEX Phenocycler platform coupled to a Zeiss Axio Observer and a Zeiss Cell Discoverer7 including an LSM900 Airy Scan confocal for increased resolution (about 100nm). The different systems currently analyze different specimen, ranging from mouse to human and covering different tissues and organs.
Contact: Andreas Schlitzer (LIMES) andreas.schlitzer@uni-bonn.de
Contact: Michael Hölzel (UKB) Michael.Hoelzel@ukbonn.de
Contact: Christian Kurts (UKB) ckurts@uni-bonn.de
Contact: Veronika Lukacs-Kornek (UKB) vlukacsk@uni-bonn.de
Contact: Alexander Pfeifer (UKB) alexander.pfeifer@uni-bonn.de
Contact: Eugenio Fava (DZNE) Eugenio.Fava@dzne.de
Deep visual proteomics
The Institute of Innate Immunity together with the newly established Translational Mass spectrometry facility at the Medical Faculty (Felix Meißner) has established a deep visual proteomics (DVP) set-up, which combines artificial-intelligence-driven image analysis of cellular phenotypes with automated single-cell or single-nucleus laser microdissection and ultra-high-sensitivity mass spectrometry. DVP links protein abundance to complex cellular or subcellular phenotypes while preserving spatial context (Mund et al., 2022 Nat. Biotechnology). The DZNE also operates a Zeiss PALM microdissection setup with a robomover with automated artificial intelligence-driven object detection-delineation-cutting-and-collection workflow.
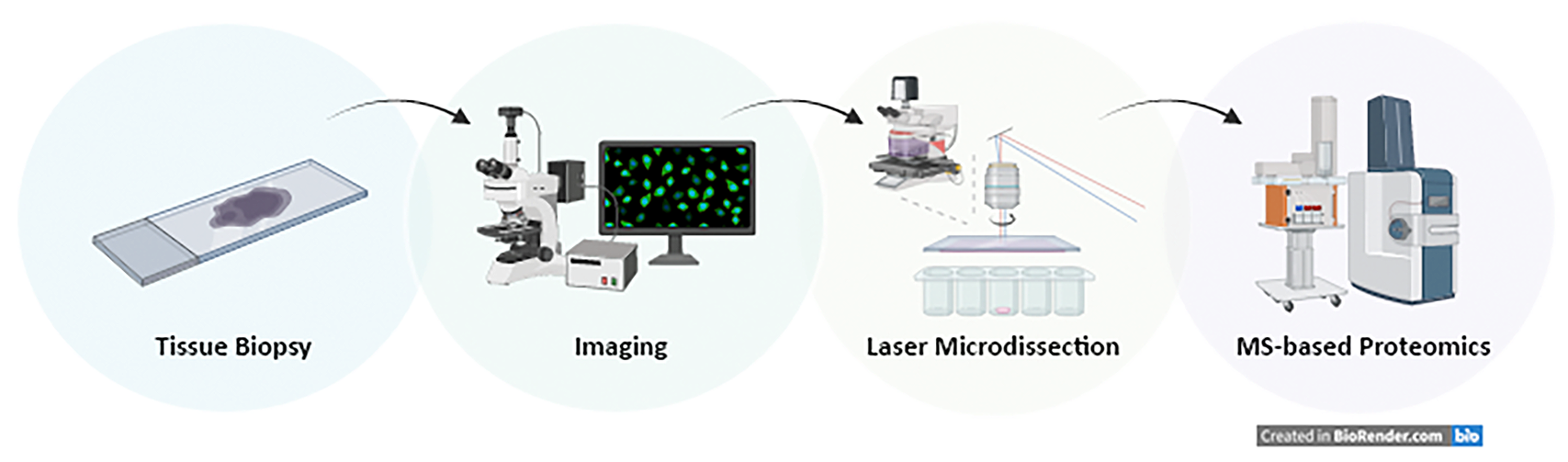
Felix Meissner (UKB) felix.meissner@uni-bonn.de
Eugenio Fava (DZNE) Eugenio.Fava@dzne.de
Steering Committee
Prof. Dagmar Wachten (UKB)
Prof. Felix Meissner (UKB)
Prof. Andreas Schlitzer (LIMES)
Prof. Michael Hölzel (UKB)
Prof. Özgün Gökce (UKB, DZNE)
Prof. Marc Beyer (DZNE)
